Application Example 1:
A part of your current lead structure is thought to be responsible for an unwanted effect in a safety assay. You plan to replace this substructure by something that is a) chemically feasible and b) likely keeping the activity against the primary target. The SwissBioisostere database might be of help in this process. By entering the substructure you would like to replace in your current compound, you will receive a list of replacements that have been performed and published in the past by medicinal chemists.In order to gain access to this information, you have to enter the substructure that you want to replace into the sketcher entitled "Fragment 1". Please mark each attachment point with an R-group (R). The SwissBioisostere database contains information on replacements for substructures with up to three attachment points.
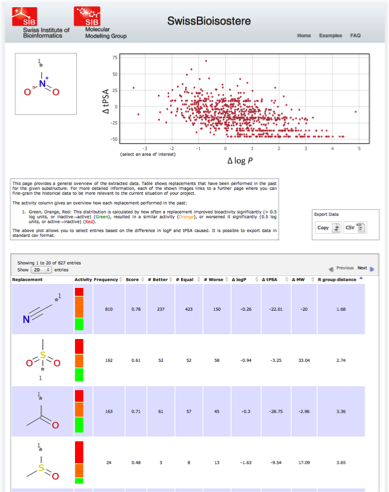
See the following screenshot as an example (click to enlarge): The time you have to wait for the query to be performed is depending on the number of replacements observed. Usually, a query should take between a second to a minute. The resulting page will give you an extensive overview about which replacements for the queried substructure have been observed in our database. You have the possibility to filter the results based on the difference in calculated logP and tPSA by selecting the range in the upper plot. The table contains additional information on e.g. observed frequency, activity difference distribution, success-based score, or chemical similarity between the fragments. All filtered results can be exported via copy to the clipboard or download of an csv-file.
See the following screenshot as an example (click to enlarge):
Application Example 2:
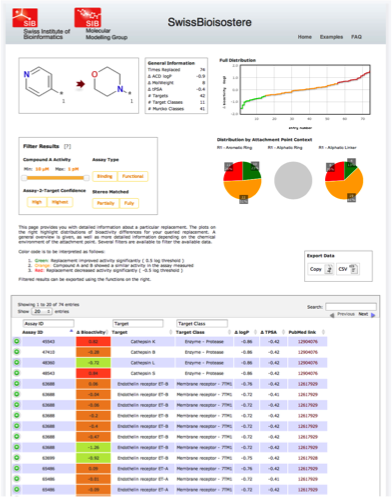
You already have an idea on how to replace a particular substructure in your molecule or are generally interested to see how a particular replacement has been observed from a bioactivity point of view. Then, you can use the SwissBioisostere to lookup information about this exact replacement. To do so, please enter the two corresponding fragments into the molecular sketchers named "Fragment 1" and "Fragment 2". Do not forget to characterize the attachment points on both fragments by R-groups R1-R3. Be aware that the given results will be sensitive to how you label these attachment points (in case a substructure is non-symmetric).See the following screenshot as an example (click to enlarge): When pressing "Query Database" you will have to wait for short time, before being presented with detailed information about this particular replacement. Besides general information being presented on the left, the right part of the screen shows you activity difference distributions for the full dataset as well as separated depending on the chemical context of the fragments. We differentiate between fragments bound to an aromatic ring, an aliphatic ring, or an aliphatic linker. These distributions are sensitive to the selection you might perform on the data. Different filter options are available. The column filter within the table provides you with the option to filter for particular target classes, targets, or assay ids. The filter box provides you with additional possibilities to fine-grain the data. You can select the range of activity of compound A of a matched pair A-B. Here, you can analyze for example whether or not a replacement showed a similar or improved bioactivity over the whole activity range or only in a particular region. Furthermore, you can split the data according to results from functional or binding assays, according to the confidence the ChEMBL curators attributed to the assay/target annotation, and if you want to consider only perfectly matched stereochemistry or also matched molecular pairs, in which stereochemistry was not completely defined for one or both of the two molecules. Compound pairs with differing stereochemistry were not considered as a matched molecular pair. All filtered results can be exported via copy to the clipboard or download of an csv-file.
See the following screenshot as an example (click to enlarge):
We hope these examples provided you with an idea of what you can expect from our database and wish you all the best for your current and future scientific endeavors.